UOMOP
Overfitting 극복 using GAP(GlobalAveragePooling) 본문
GAP(GlobalAveragePooling)은 overfitting 현상을 조금 방지할 수 있다. Flatten을 이용해서 1차원 데이터로 만든 후 모든 노드에 연결을 시켜주면 parameter가 상당히 많이 나온다. parameter가 너무 많이 나오면 해당 데이터에만 과도 학습되어 overfitting이 발생하는데, 이를 GAP을 통해서 조금 해결해보도록 한다.
GAP는 한 feature map의 하나의 channel 전체 값을 평균 내어 1개의 unit을 생성시킨다. 이 unit들을 노드에 연결시켜 parameter가 과하게 나오는 것을 막는 것이다.
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from tensorflow.keras.datasets import cifar10
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Input, Conv2D, BatchNormalization, Activation, MaxPooling2D, \
Flatten, Dropout, Dense, GlobalAveragePooling2D
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping
def make_zero_to_one(images, labels) :
images = np.array(images/255., dtype = np.float32)
labels = np.array(labels, dtype = np.float32)
return images, labels
def ohe(labels) :
labels = to_categorical(labels)
return labels
def tr_val_test(train_images, train_labels, test_images, test_labels, val_rate) :
tr_images, val_images, tr_labels, val_labels = \
train_test_split(train_images, train_labels, test_size = val_rate)
return (tr_images, tr_labels), (val_images, val_labels), (test_images, test_labels)
def create_before_model(tr_images, verbose):
input_size = tr_images.shape[1]
input_tensor = Input(shape=(input_size, input_size, 3))
x = Conv2D(filters=32, kernel_size=(3, 3), padding='same')(input_tensor)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv2D(filters=32, kernel_size=(3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = MaxPooling2D(pool_size=(2, 2))(x)
x = Conv2D(filters=64, kernel_size=3, padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv2D(filters=64, kernel_size=3, padding='same')(x)
x = Activation('relu')(x)
x = Activation('relu')(x)
x = MaxPooling2D(pool_size=2)(x)
x = Conv2D(filters=128, kernel_size=3, padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv2D(filters=128, kernel_size=3, padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Flatten(name='flatten')(x)
x = Dropout(rate=0.5)(x)
x = Dense(300, activation='relu', name='fc1')(x)
x = Dropout(rate=0.3)(x)
output = Dense(10, activation='softmax', name='output')(x)
model = Model(inputs=input_tensor, outputs=output)
return model
if verbose == True :
model.summary()
def create_after_model(tr_images, verbose):
input_size = tr_images.shape[1]
input_tensor = Input(shape=(input_size, input_size, 3))
x = Conv2D(filters=32, kernel_size=(3, 3), padding='same')(input_tensor)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv2D(filters=32, kernel_size=(3, 3), padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = MaxPooling2D(pool_size=(2, 2))(x)
x = Conv2D(filters=64, kernel_size=3, padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv2D(filters=64, kernel_size=3, padding='same')(x)
x = Activation('relu')(x)
x = Activation('relu')(x)
x = MaxPooling2D(pool_size=2)(x)
x = Conv2D(filters=128, kernel_size=3, padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv2D(filters=128, kernel_size=3, padding='same')(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = GlobalAveragePooling2D()(x)
x = Dropout(rate=0.5)(x)
x = Dense(300, activation='relu', name='fc1')(x)
x = Dropout(rate=0.3)(x)
output = Dense(10, activation='softmax', name='output')(x)
model = Model(inputs=input_tensor, outputs=output)
return model
if verbose == True :
model.summary()
def lets_compare_two(before, after) :
fig, axs = plt.subplots(nrows = 1, ncols = 2, figsize = (22, 6))
axs[0].plot(before.history["val_accuracy"], label = "before")
axs[0].plot(after.history["val_accuracy"], label = "after")
axs[0].set_title("val_accuracy")
axs[0].set_xlabel("epochs")
axs[0].set_ylabel("val_acc")
axs[0].legend()
axs[1].plot(before.history["val_loss"], label = "before")
axs[1].plot(after.history["val_loss"], label = "after")
axs[1].set_title("val_loss")
axs[1].set_xlabel("epochs")
axs[1].set_ylabel("val_loss")
axs[1].legend()
plt.show()
(train_images, train_labels), (test_images, test_labels) = cifar10.load_data()
train_images, train_labels = make_zero_to_one(train_images, train_labels)
test_images, test_labels = make_zero_to_one(test_images, test_labels)
train_labels = ohe(train_labels)
test_labels = ohe(test_labels)
(tr_images, tr_labels), (val_images, val_labels), (test_images, test_labels) = \
tr_val_test(train_images, train_labels, test_images, test_labels, val_rate = 0.15)
model_before = create_before_model(tr_images, verbose = True)
model_before.compile(optimizer = Adam(learning_rate = 0.001), loss = "categorical_crossentropy", metrics = ["accuracy"])
rlr = ReduceLROnPlateau(monitor = "val_loss", factor = 0.2, patience = 5, mode = "min", verbose = True)
ely = EarlyStopping(monitor = "val_loss", patience = 13, mode = "min", verbose = True)
result_before = model_before.fit(x = tr_images, y = tr_labels, batch_size = 32, epochs = 40, shuffle = True,
validation_data = (val_images, val_labels), callbacks = [rlr, ely])
model_after = create_after_model(tr_images, verbose = True)
model_after.compile(optimizer = Adam(learning_rate = 0.001), loss = "categorical_crossentropy", metrics = ["accuracy"])
rlr = ReduceLROnPlateau(monitor = "val_loss", factor = 0.2, patience = 5, mode = "min", verbose = True)
ely = EarlyStopping(monitor = "val_loss", patience = 13, mode = "min", verbose = True)
result_after = model_after.fit(x = tr_images, y = tr_labels, batch_size = 32, epochs = 40, shuffle = True,
validation_data = (val_images, val_labels), callbacks = [rlr, ely])
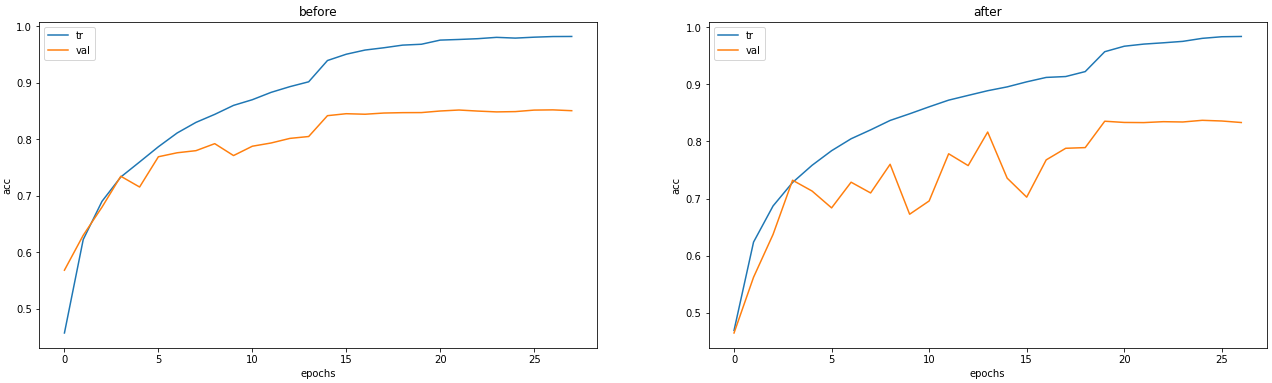
fig, axs = plt.subplots(nrows = 1, ncols = 2, figsize = (22, 6))
axs[0].plot(result_before.history["accuracy"], label = "tr")
axs[0].plot(result_before.history["val_accuracy"], label = "val")
axs[0].set_title("before")
axs[0].set_xlabel("epochs")
axs[0].set_ylabel("acc")
axs[0].legend()
axs[1].plot(result_after.history["accuracy"], label = "tr")
axs[1].plot(result_after.history["val_accuracy"], label = "val")
axs[1].set_title("after")
axs[1].set_xlabel("epochs")
axs[1].set_ylabel("acc")
axs[1].legend()
plt.show()눈에 띄는 변화를 확인할 수는 없었다. 다시 시도.

'Ai > DL' 카테고리의 다른 글
| Callback (ModelCheckpoint, ReduceLROnPlateau, EarlyStopping) (0) | 2022.02.10 |
|---|---|
| Overfitting 극복 using 가중치 규제 (0) | 2022.02.10 |
| 성능 향상 using (# of filters, depth) (0) | 2022.02.09 |
| 성능 향상 using callback (0) | 2022.02.09 |
| 성능 향상 using batch size (0) | 2022.02.09 |
Comments